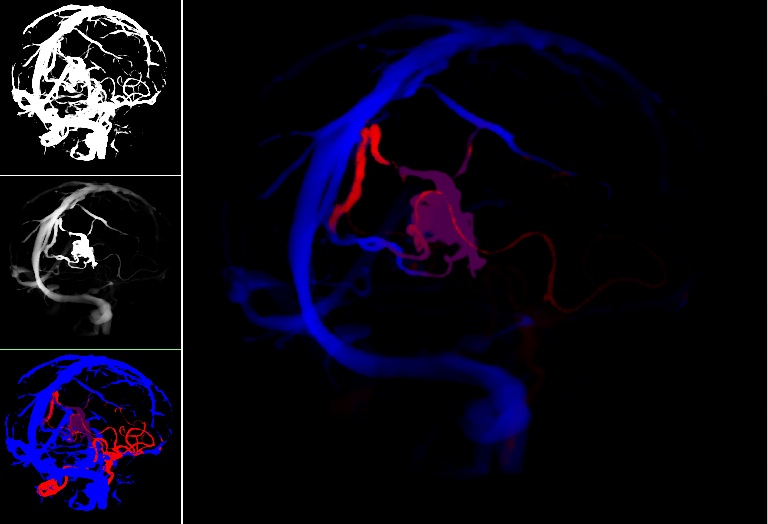
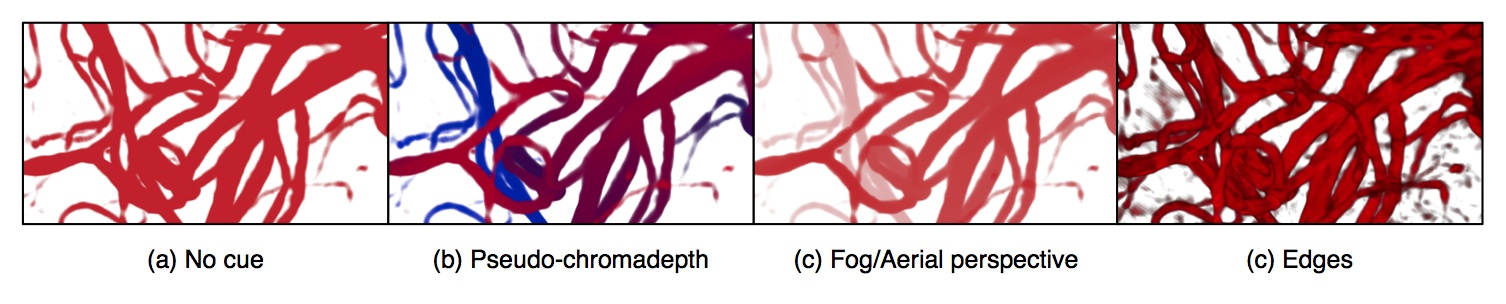
Augmented reality (AR) visualization in image-guided neurosurgery (IGNS) allows a surgeon to see rendered preoperative medical datasets (e.g. MRI/CT) from a navigation system merged with the surgical field of view. Combining the real surgical scene with the virtual anatomical models into a comprehensive visualization has the potential of reducing the cognitive burden of the surgeon by removing the need to map preoperative images and surgical plans from the navigation system to the patient. Furthermore, it allows the surgeon to see beyond the visible surface of the patient, directly at the anatomy of interest, which may not be readily visible.
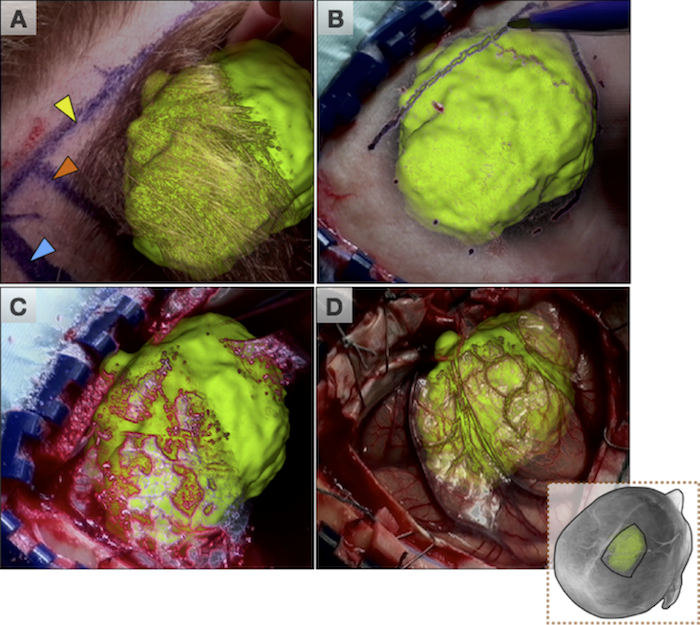
Figure: Augmented reality visualizations from our neuronavigation system. The surgeon used AR for craniotomy planning on the skin (A), the bone (B), the dura (C), and also after the craniotomy on the cortex (D). In A, the orange arrow indicated the posterior boundary of the tumour and the blue arrow indicates the planned posterior boundary of the craniotomy that will allow access to the tumour. The yellow arrow shows the medial extent of the tumour, which is also the planned craniotomy margin. In B, the surgeon uses the augmented reality view to trace around the tumour in order to determine the size of the bone flap to be removed. In C, AR is used prior to the opening of the dura and in D the tumour is visualized on the cortex prior to its resection.
Video
Publications
- M. Kersten-Oertel, I. J. Gerard , S. Drouin , J. A. Hall , D. L. Collins. “Intraoperative Craniotomy Planning for Brain Tumour Surgery using Augmented Reality”, to be presented at CARS 2016.
- I. J. Gerard, M. Kersten-Oertel, S. Drouin, J. A. Hall, K. Petrecca, D. De Nigris, T. Arbel and D. L. Collins. (2016) “Improving Patient Specific Neurosurgical Models with Intraoperative Ultrasound and Augmented Reality Visualizations in a Neuronavigation Environment,” in 4th Workshop on Clinical Image-based Procedures: Translational Research in Medical Imaging, LNCS 9401, pp. 1–8.*** Best Paper
- Kersten-Oertel, M., Gerard, I. J., Drouin, S., Mok, K., Petrecca, K., & Collins, D. L. (2015) Augmented Reality for Brain Tumour Resections. Int J CARS, 10(1):S260.