A number of unbiased non-linear averages of the MNI152 database have been generated that combines the attractions of both high-spatial resolution and signal-to-noise while not being subject to the vagaries of any single brain (Fonov et al., 2011). The procedure involved multiple iterations of a process where, at each iteration, individual native MRIs were non-linearly fitted to the average template from the previous iteration, beginning with the MNI152 linear template.
We present an unbiased standard magnetic resonance imaging template brain volume for normal population. These volumes were created using data from ICBM project.
6 different templates are available:
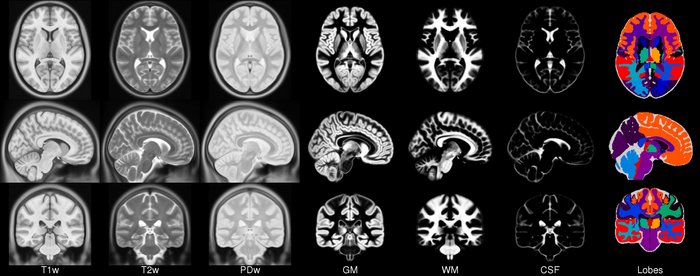
- ICBM 2009a Nonlinear Symmetric – 1×1x1mm template which includes T1w,T2w,PDw modalities, also T2 relaxometry (T2 values calculated for each subject using single dual echo PD/T2 scan), and tissue probabilities maps. Also included lobe atlas used for ANIMAL+INSECT segmentation, brain mask, eye mask and face mask. Intensity inhomogeneity was performed using N3 version 1.10.1.
- ICBM 2009a Nonlinear Asymmetric template – 1×1x1mm template which includes T1w,T2w,PDw modalities, and tissue probabilities maps. Intensity inhomogeneity was performed using N3 version 1.10.1. Also included brain mask, eye mask and face mask.
- ICBM 2009b Nonlinear Symmetric – 0.5×0.5×0.5mm template which includes only T1w,T2w and PDw modalities.
- ICBM 2009b Nonlinear Asymmetric – 0.5×0.5×0.5mm template which includes only T1w,T2w and PDw modalities.
- ICBM 2009c Nonlinear Symmetric – 1×1x1mm template which includes T1w,T2w,PDw modalities, and tissue probabilities maps. Also included lobe atlas used for ANIMAL+INSECT segmentation, brain mask, eye mask and face mask. Intensity inhomogeneity was performed using N3 version 1.11. Sampling is different from 2009a template.
- ICBM 2009c Nonlinear Asymmetric template – 1×1x1mm template which includes T1w,T2w,PDw modalities, and tissue probabilities maps. Intensity inhomogeneity was performed using N3 version 1.11 Also included brain mask, eye mask and face mask.Sampling is different from 2009a template.
All templates are describing the same anatomy, but sampling and or pre-processing is different: different versions of N3 algorithm produces slightly different tissue probability maps. Tools for using these atlases can be found in the Software section.
New: CerebrA: Mindboggle atlas adapted for ICBM 2009c template
- CerebrA labels are based on an accurate non-linear registration of cortical and subcortical labelling from Mindboggle 101 to the symmetric ICBM 2009c atlas, followed by manual editing.
Viewing
Viewing the multiple atlas volumes online requires Java browser support. The Java Internet Viewer (JIV2) used here is available for download and personal use under the GNU general public license (GPL). To view the ICBM 2009 atlases online Click “View with JIV” links below. The MNI stereotaxic coordinates (X,Y,Z) are displayed in the first row below the volumes.
For viewing, one can use the left most mouse button to click on any image, and the other cross-sectional images will be updated with the appropriate position. You can also hold the middle/rocker mouse button down while moving up or down, to pan through the image plane. Holding ‘Shift’ with the left or middle button will enable dragging and zooming.
When looking at the images, remember that left is left and right is right!
Image pre-processing included non-uniform intensity correction (Sled, 1998) and intensity normalization to a range of 0–100. All T1w MRI data was then transformed into the Talairach-like MNI stereotaxic space using minctracc (Collins, Neelin et al. 1994). Brain masking was performed using BET (Smith, 2002). Age-based subgroups of subjects were created, and all scans within each group were then automatically re-registered to the stereotaxic space using the appropriate template. For each group, an iterative nonlinear co-registration algorithm (Grabner, Janke et al. 2006, Fonov, 2011), was applied to obtain the group averages. The T1-based transformation was then applied to the T2, PD and tissue classified volumes to generate average atlases for these data. Methodological details can be found in (Fonov, 2011).
All necessary files are distributed with MINC1 and MINC2 versions of the 2009a and 2009c templates, use lobe_segment.pl script, it is also distributed as part of Minc Tool Kit
Software used to create average anatomical templates is available at Github, numerical identifiers of structures are explained in icbm2009_lobe_defs.txt
Demographics (same as ICBM152_linear)
- Handedness was scored on a scale of 0-10. Average(std dev) for 141 of the 152 subjects was 8.57(2.50) with a range of 1-10 and interquartile range 8-10.
- Age, measured in years: 25.02(4.90)y, range 18-44y, interquartile range 21-28y
- 86 males and 66 females
- Ethnic background: 129 caucasian, 15 asian and 1 mixed decent
Publications
The following publications should be referenced when using this atlas:- VS Fonov, AC Evans, K Botteron, CR Almli, RC McKinstry, DL Collins and BDCG, Unbiased average age-appropriate atlases for pediatric studies, NeuroImage,Volume 54, Issue 1, January 2011, ISSN 1053–8119, DOI: 10.1016/j.neuroimage.2010.07.033
- VS Fonov, AC Evans, RC McKinstry, CR Almli and DL Collins, Unbiased nonlinear average age-appropriate brain templates from birth to adulthood, NeuroImage, Volume 47, Supplement 1, July 2009, Page S102 Organization for Human Brain Mapping 2009 Annual Meeting, DOI: https://dx.doi.org/10.1016/S1053-8119(09)70884-5
Anatomical Structures Segmentation
- DL Collins, AP Zijdenbos, WFC Baaré and AC Evans, ANIMAL+INSECT: Improved Cortical Structure Segmentation,IPMI Lecture Notes in Computer Science, 1999, Volume 1613/1999, 210–223, DOI: https://dx.doi.org/10.1007/3-540-48714-X_16
- AL Manera; M Dadar, VS Fonov, DL Collins, CerebrA: Accurate registration and manual label correction of the Mindboggle 101 atlas for the MNI-ICBM152 template. In press
Copyright (C) 1993–2004 Louis Collins, McConnell Brain Imaging Centre, Montreal Neurological Institute, McGill University. Permission to use, copy, modify, and distribute this software and its documentation for any purpose and without fee is hereby granted, provided that the above copyright notice appear in all copies. The authors and McGill University make no representations about the suitability of this software for any purpose. It is provided “as is” without express or implied warranty. The authors are not responsible for any data loss, equipment damage, property loss, or injury to subjects or patients resulting from the use or misuse of this software package.
| ICBM 2009a Nonlinear Symmetric 1×1x1mm template: MINC2 72MB MINC1 75MB NIFTI 56MB View with JIV |
 |
| ICBM 2009a Nonlinear Asymmetric 1×1x1mm template: MINC2 58MB MINC1 58MB NIFTI 57MB |
 |
 |
| ICBM 2009b Nonlinear Asymmetric 0.5×0.5×0.5mm template: MINC2 366MB MINC1 367MB NIFTI 358MB |
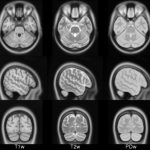 |
| ICBM 2009c Nonlinear Symmetric 1×1x1mm template: MINC2 57MB MINC1 57MB NIFTI 55MB View with JIV |
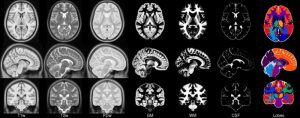 |
| ICBM 2009c Nonlinear Asymmetric 1×1x1mm template: MINC2 57MB MINC1 57MB NIFTI 57MB |
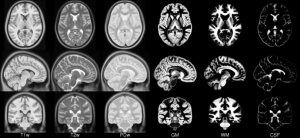 |
| CerebrA atlas MINC2 MINC1 NIFTI |
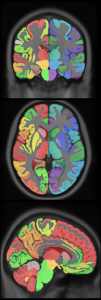 |
For questions related to the MNI ICBM 2009 atlases (rather than the website), contact Vladimir Fonov