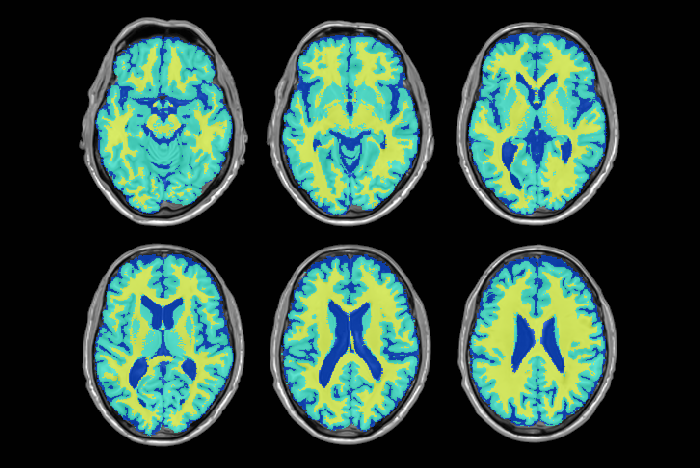
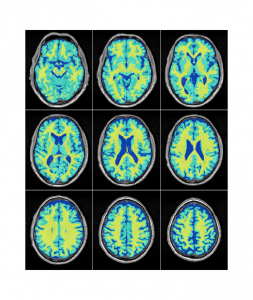
Accurate differentiation of brain tissue types from MR images is necessary in many neuroscience and clinical applications. Accurate automated tissue segmentation is challenging due to the variability in the tissue intensity profiles caused by differences in scanner models, acquisition protocols, as well as the age of the subjects and presence of pathology. We have developed BISON (Brain tIsue SegmentatiOn pipeliNe), a new pipeline for tissue segmentation using a random forests classifier and a set of intensity and location priors obtained based on T1w images.
Execution Example:
python BISON.py -c RF -m Trained_Classifiers/ -o Outputs/ -t Temp_Files/ -e PT -n List.csv -p Trained_Classifiers/ -l 3